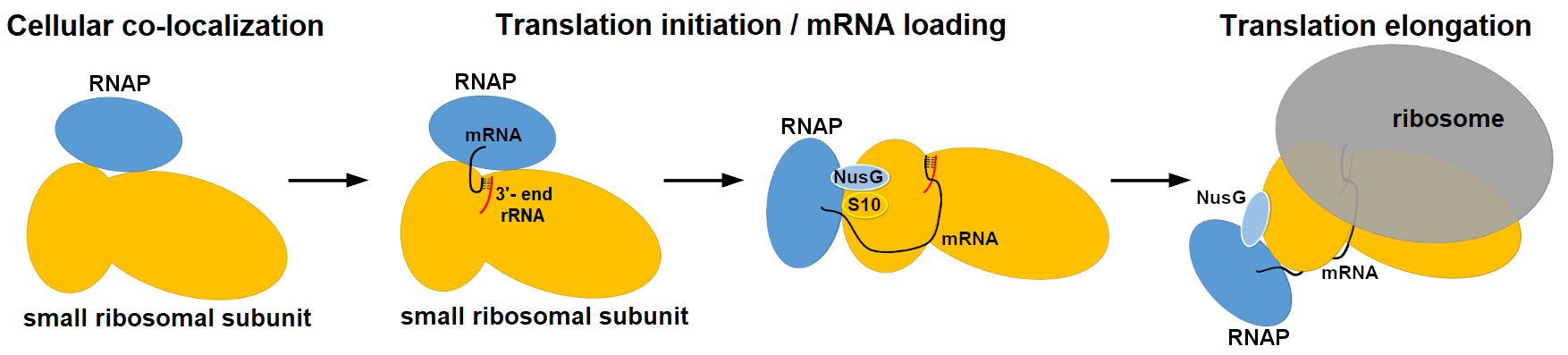
In Bacteria and Archaea, newly transcribed RNAs are immediately bound by ribosomes, thus coupling transcription to translation. Indeed, recent cryo-electron microscopy (cryo-EM) studies report that RNA polymerase (RNAP) and ribosome may interact directly or with the help of transcription factors. This contrasts with eukaryotic transcription and translation which are physically separated by the nuclear envelope. Nevertheless, some double-strand DNA viruses replicate in cytoplasmic factories of infected eukaryotic cells, raising the possibility that viral transcription might be directly coupled to translation by host ribosomes in vivo.
To probe the mechanistic details and functional outcomes of coupled transcription-translation, we use an integrative approach that combines proteomics, molecular biology, biochemistry and structural biology. We aim to characterize transcription-translation coupling in bacteria (E. Coli) and virus-infected cells in the context of ribosome interactions with RNAP, DNA, mRNA, and transcription or translation factors. The fundamental strength of our approach lies in the use of time-resolved cryo-EM and cryo-electron tomography (cryo-ET) that enable us to characterize various points of transcriptional-translational apparatus in bacteria and virus infected mammalian cells from the atomic resolution of individually reconstituted macromolecules in vitro (cryo-EM) to the supramolecular complexes in vivo (cryo-ET) within thin sections of bacterial or infected cells in near native conditions.
These studies help to understand how the leading translating ribosomes preserve genome integrity by preventing RNAP from backtracking or pausing in bacteria, and whether an initial round of translation in the nucleoid guarantees the message for steady translation in cytoplasm. Moreover, detailed structural information about coupled systems in virus-infected cells can bring crucial insights into the mechanisms of viral pathogenesis in late stages of viral infection.