Bacterial transcription-translation coupling
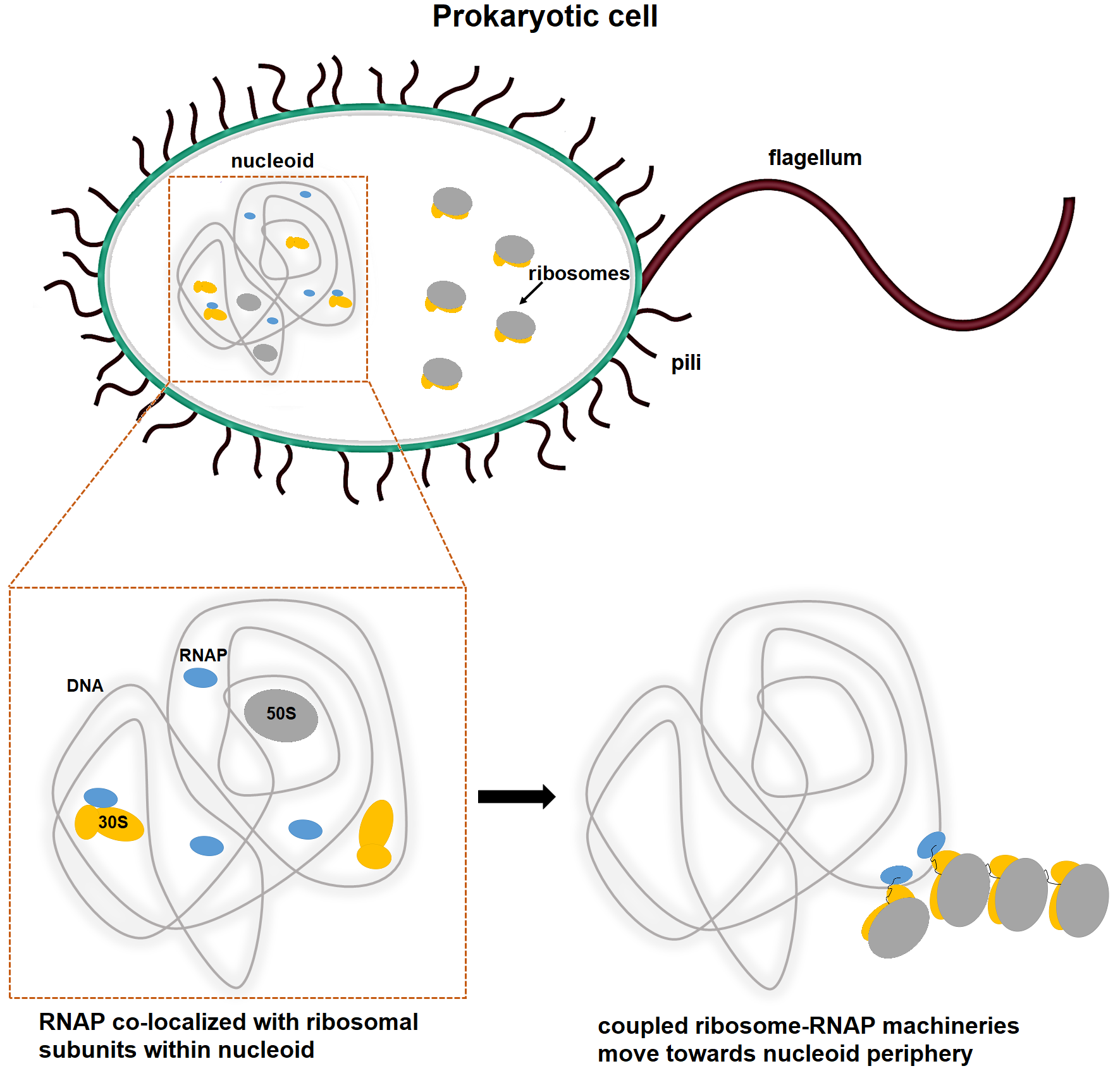
In prokaryotic cell, RNA polymerases (RNAP) and free ribosome subunits can explore the entire nucleoid (lower left panel). RNAP starts transcribing a gene within the nucleoid and the first ribosome binds to the emerging RNA. As the polyribosomal structures grow, the coupled transcription-translation apparatus possibly moves to the periphery of the nucleoid (lower right panel).
We use time resolved single-particle cryo-electron microscopy (cryo-EM) to determine high resolution structures of in vitro RNAP-ribosome complexes at multiple states of coupled transcription-translation: from translation initiation to elongation. The structures of in vitro complexes will be compared to in vivo complexes visualized by cryo-electron tomography (cryo-ET) at the near-nucleoid space.
Viral transcription-translation coupling in mammalian cells
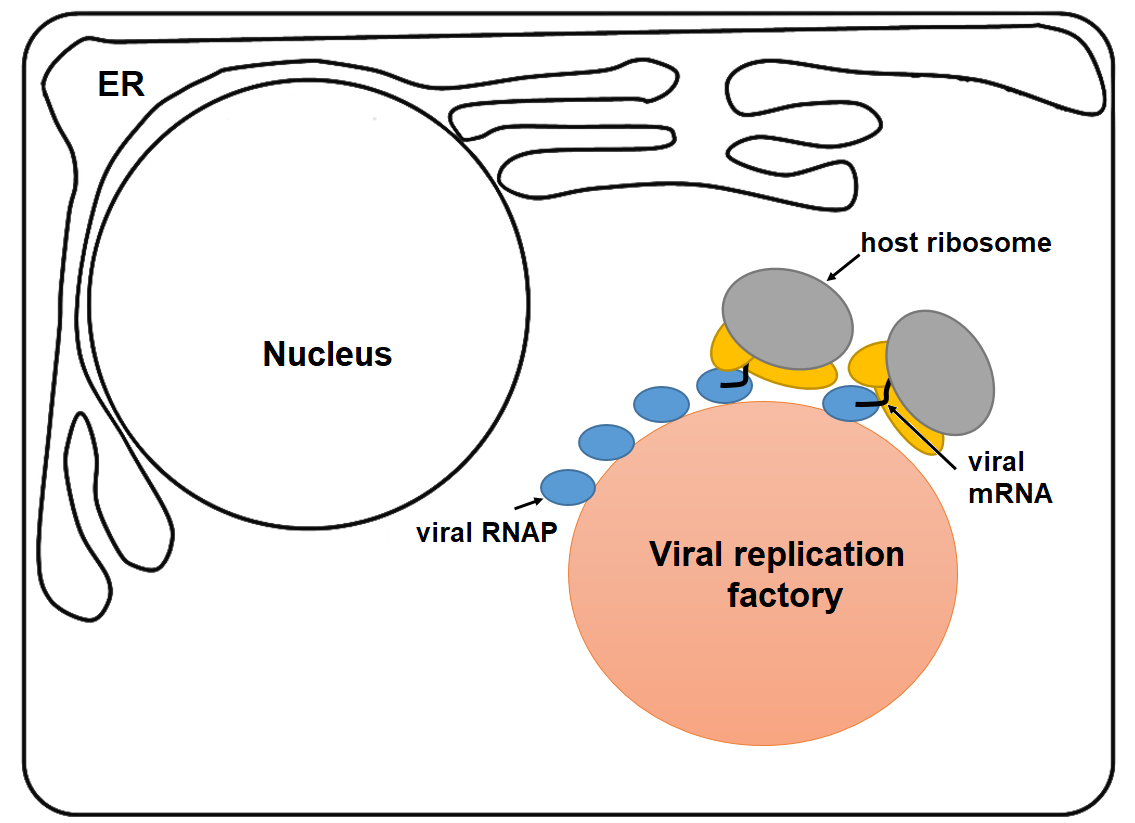
A simplified diagram of transcription-translation coupling in infected mammalian cell by cytoplasmic virus at late stages of viral infection. The transcribed viral mRNAs by viral RNAPs concentrate in viral factories and closely associate with ribosomes to maximize the production of viral proteins in late stages of infection.
We apply cryo-ET of infected cells and apply sub-tomographic averaging to visualize viral RNAP and ribosomes at sub-nanometer resolution and determine whether and how viruses couple transcription and translation in viral factories.
Demo Lab




